Home News Technical articles How to Choose Filtration in the Pharmaceutical Industry: Microfiltration (MF) Ultrafiltration (UF) or Nanofiltration (NF)?
How to Choose Filtration in the Pharmaceutical Industry: Microfiltration (MF) Ultrafiltration (UF) or Nanofiltration (NF)?
2025-07-10 Eternalwater 1、What needs to be separated/removed? (Bacteria? Proteins? Pyrogens? Salts?)2、What is the size/molecular weight (MW) of the target substance?
3、What is the process objective? (Sterilization? Concentration? Purification? Desalting?)
There are differences in the applications of the three major membrane separation technologies: microfiltration (MF), ultrafiltration (UF), and nanofiltration (NF). It is not a simple order of "large, medium and small", but each has its unique separation principles and targets. Their core differences and key parameters - pore size and molecular weight cut-off. Understanding pore size, molecular weight cut-off and application logic is the first step to ensuring the efficiency of pharmaceutical processes and product safety.

Microfiltration(MF):

Ultrafiltration(UF):
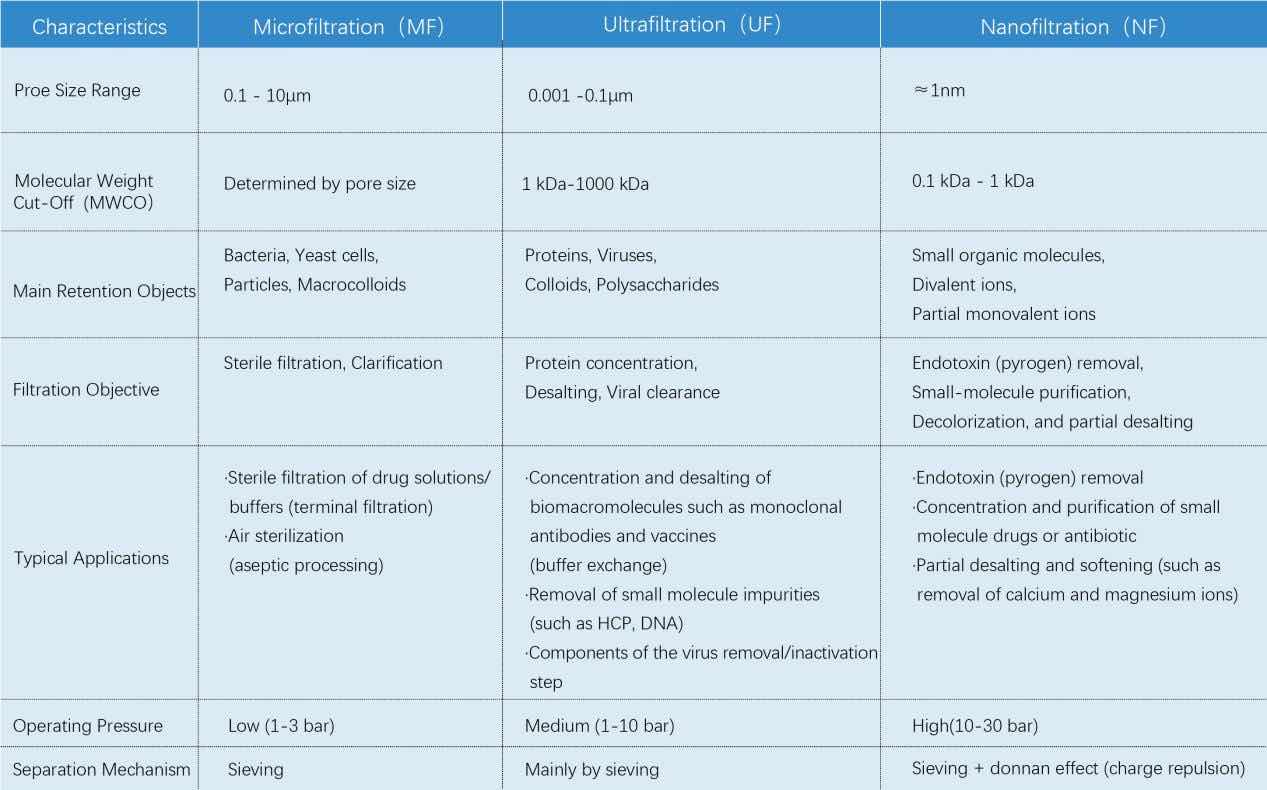
Important Notice: The Molecular Weight Cut-Off (MWCO) is a laboratory reference value! Actual separation performance is influenced by pH, ionic strength, membrane material, and other factors. Process validation is mandatory!
Macromolecules
1、Macromolecules refer to biological substances with a relative molecular mass of more than 5,000 or even more than one million, such as proteins, nucleic acids, polysaccharides, etc. These biomolecules are extremely closely related to life activities and are composed of of simple molecular units that are considered monomers. In solution, they demonstrate gel-forming capabilities. Generally, compounds with a relative molecular mass of more than 10,000 are called macromolecular compounds or polymer compounds.These substances consist of many repetitive
structural units, generally with a linear structure and some with a branch-like structure. Many substances with important biological effects, such as proteins and nucleic acids, belong to this type of compound. The basic building-block molecule of a macromolecular protein is amino acid.
Small molecules
2、Small molecules are defined as molecules with a molecular weight below 500 , which typically exist as simple monomeric substances.
From a chemical perspective, small molecules refer to biologically functional compounds with a molecular weight below 1,000 Daltons (Da), particularly those <400 Da. From a biological perspective, small molecules encompass bioactive small peptides, oligopeptides, oligosaccharides, oligonucleotides, vitamins, minerals, plant secondary metabolites and heir degradation products.
Bubble Point Test Method
3、Bubble Point Test Method:Measures the maximum pore size of the membrane - when pressure reaches the bubble point, the first bubble breakthrough corresponds to the largest pore.
Electron Microscopy:Scans the membrane surface to generate a pore size distribution map. A narrower peak indicates more uniform pore sizes.
Molecular Weight Cut-Off (MWCO)
4、A membrane retains more than 90% of the molecular weight of the specific standard substance (typically globular protein or [PEG] polyethylene glycol). The smaller the pore size, the smaller the molecular weight cut-off (Ultrafiltration (UF) Membrane Molecular Weight Cut-Off :1 kDa,3 kDa, 10 kDa, 30 kDa, 300 kDa...;Nanofiltration (NF) Membrane Molecular Weight Cut-Off:100 - 1000 Da (0.1 - 1 kDa) 。 Usually ranges between 100 - 1000 Da (0.1 - 1 kDa).
However, molecular shape has a significant impact:
·Linear molecules (such as DNA, PEG) may penetrate membranes with a lower nominal molecular weight cutoff.
·Globular/rigid molecules (such as proteins) are more likely to be retained by membranes that are close to molecular weight cut-off.
Dalton
5、Dalton, symbols: Da, Dal, or D, is the standard unit for expressing molecular weight, calculated as the sum of atomic masses of all atoms in a molecule.
"D" or "KD" are commonly used in biochemistry, molecular biology, and proteomics. Since proteins are macromolecules, their molecular weights are typically expressed in kilodaltons (kDa). These units cannot be directly converted to millimeters (mm) as they represent fundamentally different physical quantities - mass and length.
1 mDa = 1000 kDa
1 kDa(kilodalton) = 1000 Da(Dalton, Da)
1 Da ≈ 1 g/mol
Example: The molecular weight of a water molecule (H₂O) is approximately 18 Da, equivalent to 0.018 kDa.
- Industry Application
- Life Sciences
- water treatment
- Industrial Filtration
- Food & Beverage
- Microelectronics
- Laboratory
- New energy battery
- Contact Us
- [email protected]
- +86-571-87022016
- +86-571-87293027
- +8613675899519
- Subscribe for Join Us!
- Join us and get detail information,technical parameter and new products etc.
- [email protected]
- Jenny wu
- +8613675899519
- +86(571) 87022016

 EN
EN  ES
ES AR
AR JP
JP CN
CN